This section covers the spatial modeling for australian plague locusts
- section 1: data management
- section 2: outbreak modeling
- section 3: nil-observatiom modeling
- section 4: outbreak and mean annual preciptation modeling
We will do basic modeling and saving relevant data to disk
Another notebook later in the series will create mansucript figures and tables
Section 1 - Data management
load_packages <- function(packages) {
# Check for uninstalled packages
uninstalled <- packages[!packages %in% installed.packages()[,"Package"]]
# Install uninstalled packages
if(length(uninstalled)) install.packages(uninstalled, dependencies = TRUE)
# Load all packages
for (pkg in packages) {
if (!require(pkg, character.only = TRUE, quietly = TRUE)) {
message(paste("Failed to load package:", pkg))
}
}
}
# List of packages to check, install, and load
packages <- c("mgcv", "gratia", "sf","rnaturalearth",
"ggpubr", "patchwork", "broom", "knitr", "janitor",
"here","ggpubr","MetBrewer","GGally","tidyverse")
load_packages(packages)
i_am('README.md')
# Setting R options for jupyterlab
options(repr.plot.width = 10, repr.plot.height = 10,repr.matrix.max.rows=10)
here() starts at /home/datascience/herbivore_nutrient_interactions
read in data
- filter species for APL
- manage data into long format
- select variables used for modeling
- create outbreak and nil observation datasets
apl_dat <- read_csv(here('data/processed/spatial_modeling/apl_survey_remote_sense_data.csv'),show_col_types = FALSE)
apl_locust_dat <- apl_dat |>
clean_names() |>
mutate(adult_density = str_remove_all(adult_density, "[{}]"),
nymph_density = str_remove_all(nymph_density, "[{}]")) %>%
separate_rows(adult_density, sep = ", ") %>%
separate(adult_density, into = c("adult_density", "adult_density_count"), sep = ": ") %>%
separate_rows(nymph_density, sep = ", ") %>%
separate(nymph_density, into = c("nymph_density", "nymph_density_count"), sep = ": ") %>%
mutate(across(c(adult_density, adult_density_count, nymph_density, nymph_density_count), as.numeric)) |>
distinct() |>
group_by(polygon_id,species) |>
mutate(total_observations = n())
unique(apl_locust_dat$ecoregion)
- 'Great Victoria desert'
- 'Central Ranges xeric scrub'
- 'Tirari-Sturt stony desert'
- 'Simpson desert'
- 'Great Sandy-Tanami desert'
- 'Mitchell Grass Downs'
- 'Carpentaria tropical savanna'
- 'Flinders-Lofty montane woodlands'
- 'Murray-Darling woodlands and mallee'
- 'Southeast Australia temperate savanna'
- 'Naracoorte woodlands'
- 'Southeast Australia temperate forests'
- 'Eastern Australia mulga shrublands'
- 'Brigalow tropical savanna'
- 'Eastern Australian temperate forests'
- 'Eyre and York mallee'
- 'unnamed_0'
- 'species'
- 'polygon_id'
- 'date'
- 'longitude'
- 'latitude'
- 'data_quality'
- 'nymph_density'
- 'nymph_density_count'
- 'adult_density'
- 'adult_density_count'
- 'data_quality_total_count'
- 'adult_density_total_count'
- 'nymph_density_total_count'
- 'geometry_x'
- 'x0_bdod_median'
- 'x10_silt_median'
- 'x1_cec_median'
- 'x2_cfvo_median'
- 'x3_clay_median'
- 'x4_nitrogen_median'
- 'x5_ocd_median'
- 'x6_ocs_single_band'
- 'x7_phh2o_median'
- 'x8_sand_median'
- 'x9_soc_median'
- 'awc_awc_000_005_ev'
- 'awc_awc_005_015_ev'
- 'bdw_bdw_000_005_ev'
- 'bdw_bdw_005_015_ev'
- 'cly_cly_000_005_ev'
- 'cly_cly_005_015_ev'
- 'ece_ece_000_005_ev'
- 'ece_ece_005_015_ev'
- 'nto_nto_000_005_ev'
- 'nto_nto_005_015_ev'
- 'pto_pto_000_005_ev'
- 'pto_pto_005_015_ev'
- 'slt_slt_000_005_ev'
- 'slt_slt_005_015_ev'
- 'snd_snd_000_005_ev'
- 'snd_snd_005_015_ev'
- 'soc_soc_000_005_ev'
- 'soc_soc_005_015_ev'
- 'bio01'
- 'bio02'
- 'bio03'
- 'bio04'
- 'bio05'
- 'bio06'
- 'bio07'
- 'bio08'
- 'bio09'
- 'bio10'
- 'bio11'
- 'bio12'
- 'bio13'
- 'bio14'
- 'bio15'
- 'bio16'
- 'bio17'
- 'bio18'
- 'bio19'
- 'elevation'
- 'p_hc_p_hc_000_005_ev'
- 'p_hc_p_hc_005_015_ev'
- 'tree_canopy_cover'
- 'ecoregion'
- 'geometry_y'
- 'total_observations'
apl_outbreak_dat <- apl_locust_dat |>
ungroup() |>
filter(species == 11 & nymph_density == 4) |>
select(polygon_id,longitude,latitude,nymph_density,nymph_density_count,nymph_density_total_count,
nto_nto_000_005_ev,nto_nto_005_015_ev,pto_pto_005_015_ev,pto_pto_000_005_ev,ecoregion,bio12) |>
group_by(polygon_id) |>
mutate(nitrogen = sum(nto_nto_005_015_ev,nto_nto_000_005_ev)/2,
phosphorus = sum(pto_pto_005_015_ev,pto_pto_000_005_ev)/2,
ecoregion = factor(ecoregion)) |>
select(!c(nto_nto_000_005_ev,nto_nto_005_015_ev,pto_pto_005_015_ev,pto_pto_000_005_ev)) |>
distinct()
apl_nil_dat <- apl_locust_dat |>
ungroup() |>
filter(species == 11 & nymph_density == 0) |>
select(polygon_id,longitude,latitude,nymph_density,nymph_density_count,nymph_density_total_count,
nto_nto_000_005_ev,nto_nto_005_015_ev,pto_pto_005_015_ev,pto_pto_000_005_ev,ecoregion,bio12) |>
group_by(polygon_id) |>
mutate(nitrogen = sum(nto_nto_005_015_ev,nto_nto_000_005_ev)/2,
phosphorus = sum(pto_pto_005_015_ev,pto_pto_000_005_ev)/2,
ecoregion = factor(ecoregion)) |>
select(!c(nto_nto_000_005_ev,nto_nto_005_015_ev,pto_pto_005_015_ev,pto_pto_000_005_ev)) |>
distinct()
save to disk for further use
write.csv(apl_outbreak_dat,here('data/processed/spatial_modeling/spatial_modeling_locust_outbreak_model_data.csv'),row.names=FALSE)
write.csv(apl_nil_dat,here('data/processed/spatial_modeling/spatial_modeling_locust_nil_model_data.csv'),row.names=FALSE)
Section 2 - outbreak modeling
outbreak_data <- read_csv(here('data/processed/spatial_modeling/spatial_modeling_locust_outbreak_model_data.csv'),
show_col_types = FALSE) |>
mutate(ecoregion = factor(ecoregion),nymph_density_count=as.integer(nymph_density_count)) |>
#Filtering out extreme values
filter(nymph_density_count <= 50 & nitrogen <= 0.4) |>
drop_na()
head(outbreak_data,1)
nrow(outbreak_data)
A tibble: 1 × 10
| <dbl> |
<dbl> |
<dbl> |
<dbl> |
<int> |
<dbl> |
<fct> |
<dbl> |
<dbl> |
<dbl> |
| 162921 |
137.1195 |
-33.36002 |
4 |
1 |
1 |
Eyre and York mallee |
331 |
0.07035461 |
0.02197414 |
6618
aus <- ne_states(country = 'Australia')
map_dat <- outbreak_data |>
select(polygon_id,latitude,longitude,nymph_density_count) |>
st_as_sf(coords = c('longitude','latitude'),
crs= "+proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0")
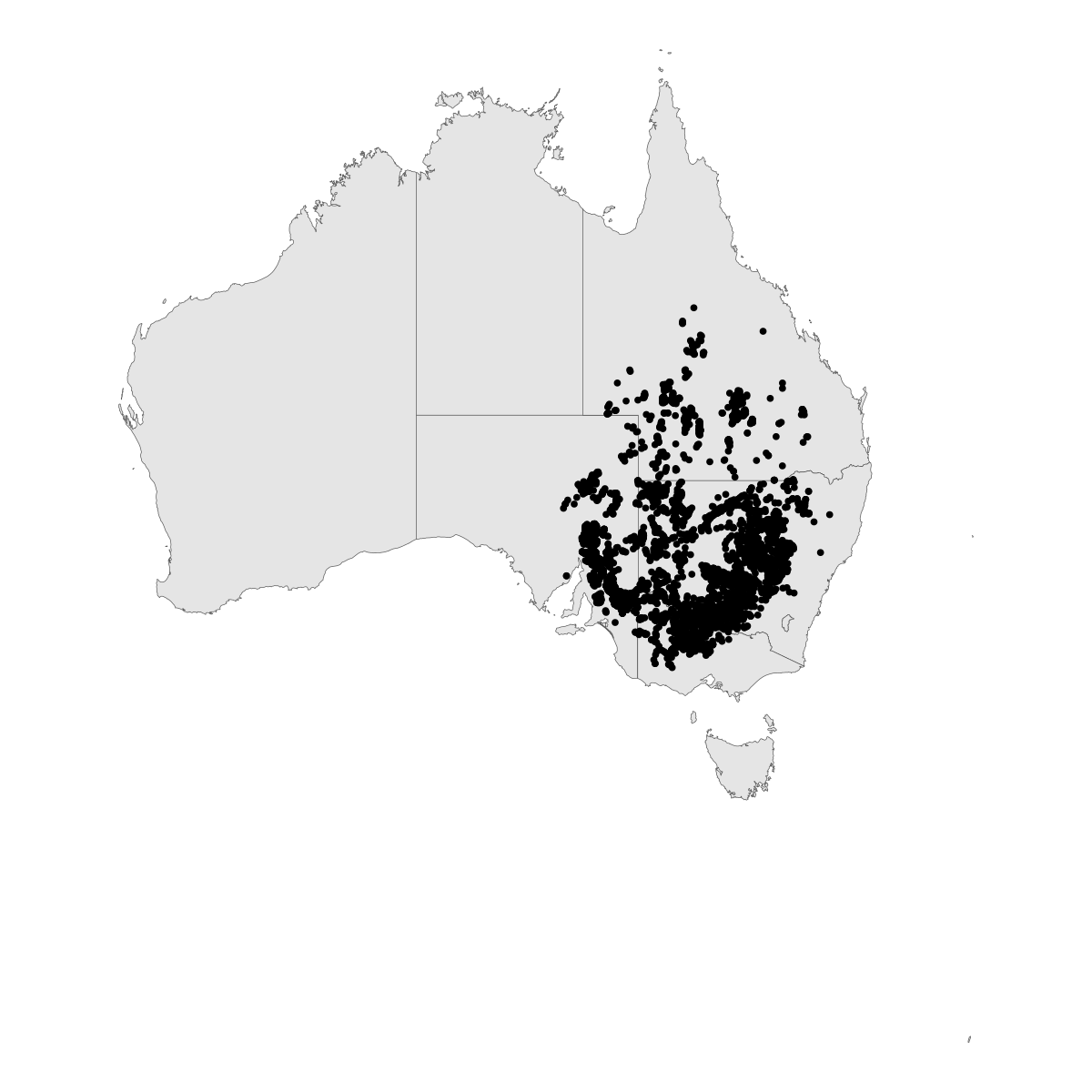
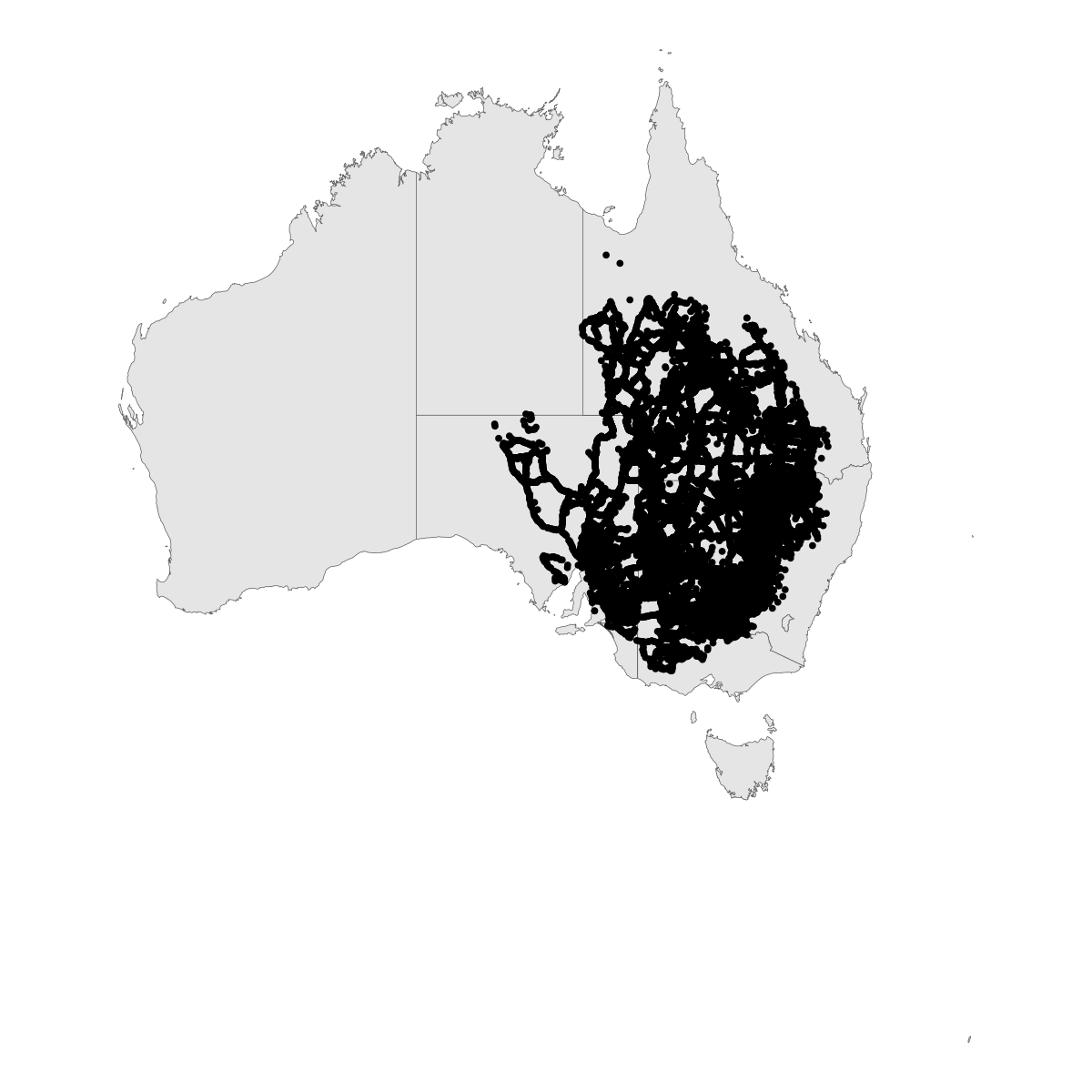
outbreak_point_map <- aus |>
ggplot() +
geom_sf() +
geom_sf(data=map_dat) +
theme_void()
outbreak_point_map
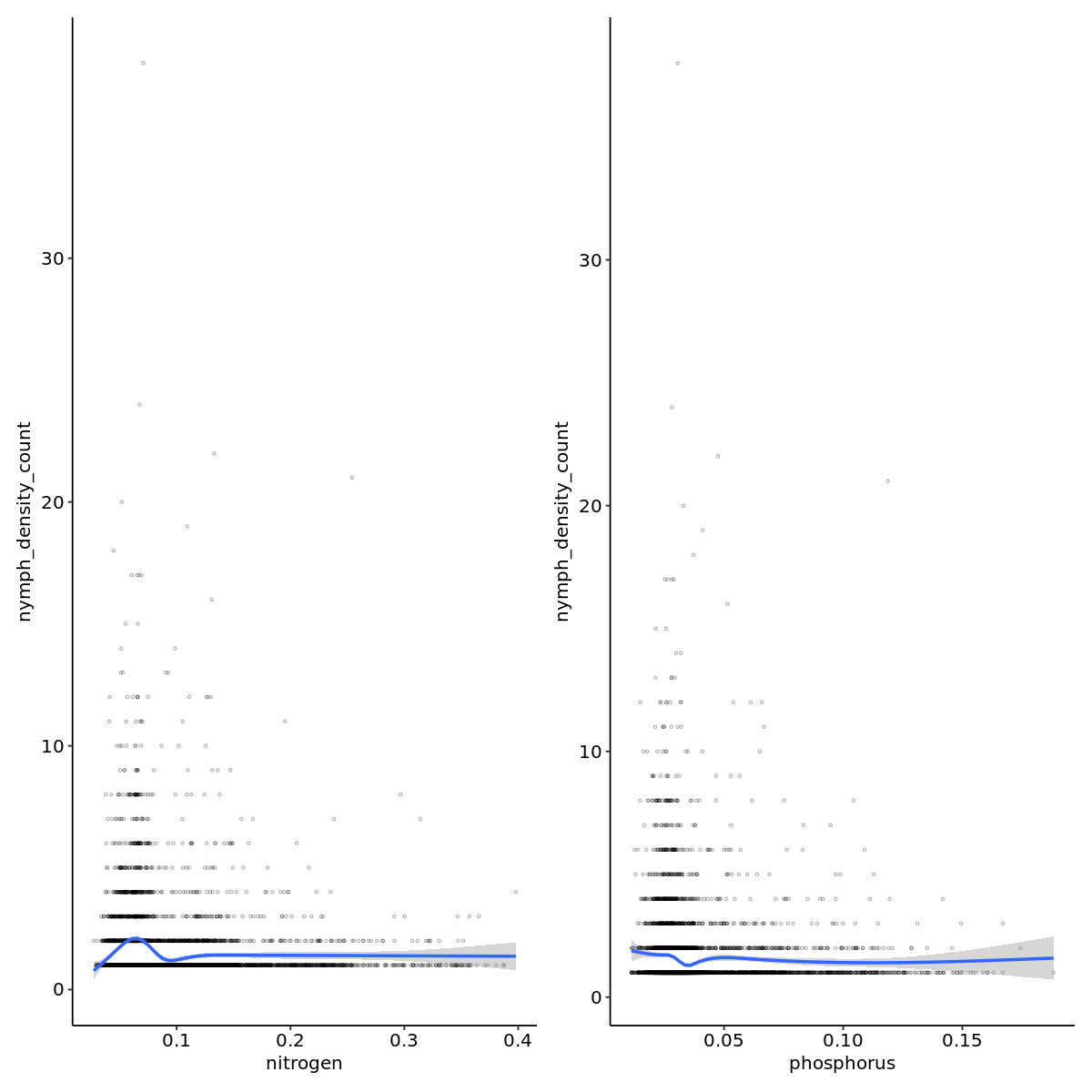
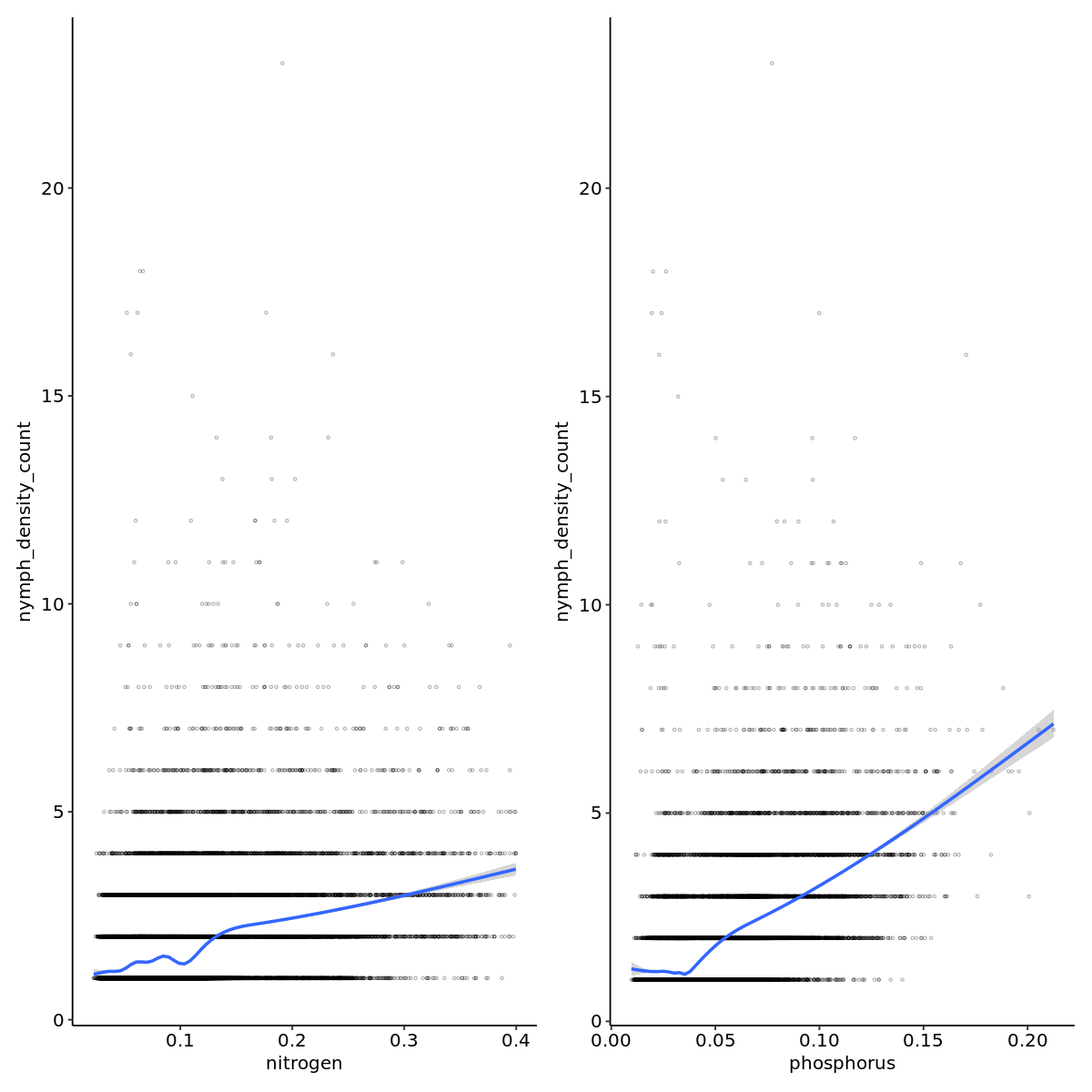
nto_plot <- outbreak_data |>
ggplot(aes(x=nitrogen,y=nymph_density_count)) +
geom_point(pch=21,alpha=0.3,size=0.6) +
geom_smooth() +
theme_pubr()
pto_plot <- outbreak_data |>
ggplot(aes(x=phosphorus,y=nymph_density_count)) +
geom_point(pch=21,alpha=0.3,size=0.6) +
geom_smooth() +
theme_pubr()
nto_plot + pto_plot
`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
polygon_id longitude latitude nymph_density
Min. : 162921 Min. :137.0 Min. :-37.57 Min. :4
1st Qu.: 437649 1st Qu.:139.7 1st Qu.:-34.45 1st Qu.:4
Median :1009080 Median :143.0 Median :-32.53 Median :4
Mean : 984545 Mean :142.9 Mean :-32.25 Mean :4
3rd Qu.:1494243 3rd Qu.:146.0 3rd Qu.:-31.04 3rd Qu.:4
Max. :2092645 Max. :151.3 Max. :-21.08 Max. :4
nymph_density_count nymph_density_total_count
Min. : 1.000 Min. : 1.000
1st Qu.: 1.000 1st Qu.: 1.000
Median : 1.000 Median : 1.000
Mean : 1.555 Mean : 2.382
3rd Qu.: 1.000 3rd Qu.: 2.000
Max. :38.000 Max. :205.000
ecoregion nitrogen
Southeast Australia temperate savanna:1969 Min. :0.02707
Murray-Darling woodlands and mallee :1407 1st Qu.:0.05658
Flinders-Lofty montane woodlands : 954 Median :0.07917
Tirari-Sturt stony desert : 584 Mean :0.09425
Southeast Australia temperate forests: 508 3rd Qu.:0.11560
Simpson desert : 469 Max. :0.61003
(Other) : 754
phosphorus
Min. :0.01107
1st Qu.:0.02562
Median :0.03093
Mean :0.03701
3rd Qu.:0.03702
Max. :0.23102
outbreak_mod <- bam(nymph_density_count ~
s(nitrogen,k=10,bs='tp') +
s(phosphorus,k=10,bs='tp') +
s(nymph_density_total_count,k=25,bs='tp') +
te(longitude,latitude,bs=c('gp','gp'),k=25) +
s(ecoregion,bs='re'),
select = TRUE,
discrete = TRUE,
nthreads = 20,
family = tw(),
data = outbreak_data)
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Family: Tweedie(p=1.99)
Link function: log
Formula:
nymph_density_count ~ s(nitrogen, k = 10, bs = "tp") + s(phosphorus,
k = 10, bs = "tp") + s(nymph_density_total_count, k = 25,
bs = "tp") + te(longitude, latitude, bs = c("gp", "gp"),
k = 25) + s(ecoregion, bs = "re")
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.55154 0.03495 44.39 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(nitrogen) 6.273 9 25.620 <2e-16 ***
s(phosphorus) 5.372 9 15.521 <2e-16 ***
s(nymph_density_total_count) 22.547 24 630.896 <2e-16 ***
te(longitude,latitude) 56.140 529 1.148 0.0123 *
s(ecoregion) 6.498 10 4.802 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
R-sq.(adj) = 0.769 Deviance explained = 83.2%
fREML = 573.15 Scale est. = 0.064377 n = 6618
A matrix: 5 × 4 of type dbl
| s(nitrogen) |
9 |
6.273422 |
0.9868934 |
0.300 |
| s(phosphorus) |
9 |
5.372169 |
1.0170037 |
0.955 |
| s(nymph_density_total_count) |
24 |
22.547306 |
0.9573451 |
0.000 |
| te(longitude,latitude) |
624 |
56.140461 |
0.8768059 |
0.000 |
| s(ecoregion) |
11 |
6.498451 |
NA |
NA |
concurvity(outbreak_mod,full=FALSE)
- $worst
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.6883534 |
0.7490074 |
0.9880373 |
1.0000000 |
1.0000000 |
| s(nitrogen) |
0.6883534 |
1.0000000 |
0.8558808 |
0.7209855 |
0.8334426 |
0.7424998 |
| s(phosphorus) |
0.7490074 |
0.8558808 |
1.0000000 |
0.7990995 |
0.8208353 |
0.7555303 |
| s(nymph_density_total_count) |
0.9880373 |
0.7209855 |
0.7990995 |
1.0000000 |
0.9890154 |
0.9883096 |
| te(longitude,latitude) |
1.0000000 |
0.8334425 |
0.8208353 |
0.9890154 |
1.0000000 |
1.0000000 |
| s(ecoregion) |
1.0000000 |
0.7424998 |
0.7555303 |
0.9883096 |
1.0000000 |
1.0000000 |
- $observed
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.5276437 |
0.7365579 |
0.9079395 |
0.01051500 |
0.01358394 |
| s(nitrogen) |
0.6883534 |
1.0000000 |
0.8458158 |
0.7027435 |
0.53716791 |
0.09132533 |
| s(phosphorus) |
0.7490074 |
0.7118931 |
1.0000000 |
0.7820410 |
0.03677412 |
0.04454272 |
| s(nymph_density_total_count) |
0.9880373 |
0.5505898 |
0.7909006 |
1.0000000 |
0.05303951 |
0.05005853 |
| te(longitude,latitude) |
1.0000000 |
0.8260141 |
0.7960190 |
0.9416384 |
1.00000000 |
0.93915689 |
| s(ecoregion) |
1.0000000 |
0.7322901 |
0.7418221 |
0.9249175 |
0.73876773 |
1.00000000 |
- $estimate
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.5552027 |
0.6733253 |
0.9666292 |
0.006421152 |
0.1758807 |
| s(nitrogen) |
0.6883534 |
1.0000000 |
0.8020399 |
0.6832916 |
0.022627257 |
0.2610817 |
| s(phosphorus) |
0.7490074 |
0.7346361 |
1.0000000 |
0.7501648 |
0.012278002 |
0.1806277 |
| s(nymph_density_total_count) |
0.9880373 |
0.5723728 |
0.7178054 |
1.0000000 |
0.010464633 |
0.2062077 |
| te(longitude,latitude) |
1.0000000 |
0.7761568 |
0.7797759 |
0.9722508 |
1.000000000 |
0.9434299 |
| s(ecoregion) |
1.0000000 |
0.6883744 |
0.6950790 |
0.9693498 |
0.040977785 |
1.0000000 |
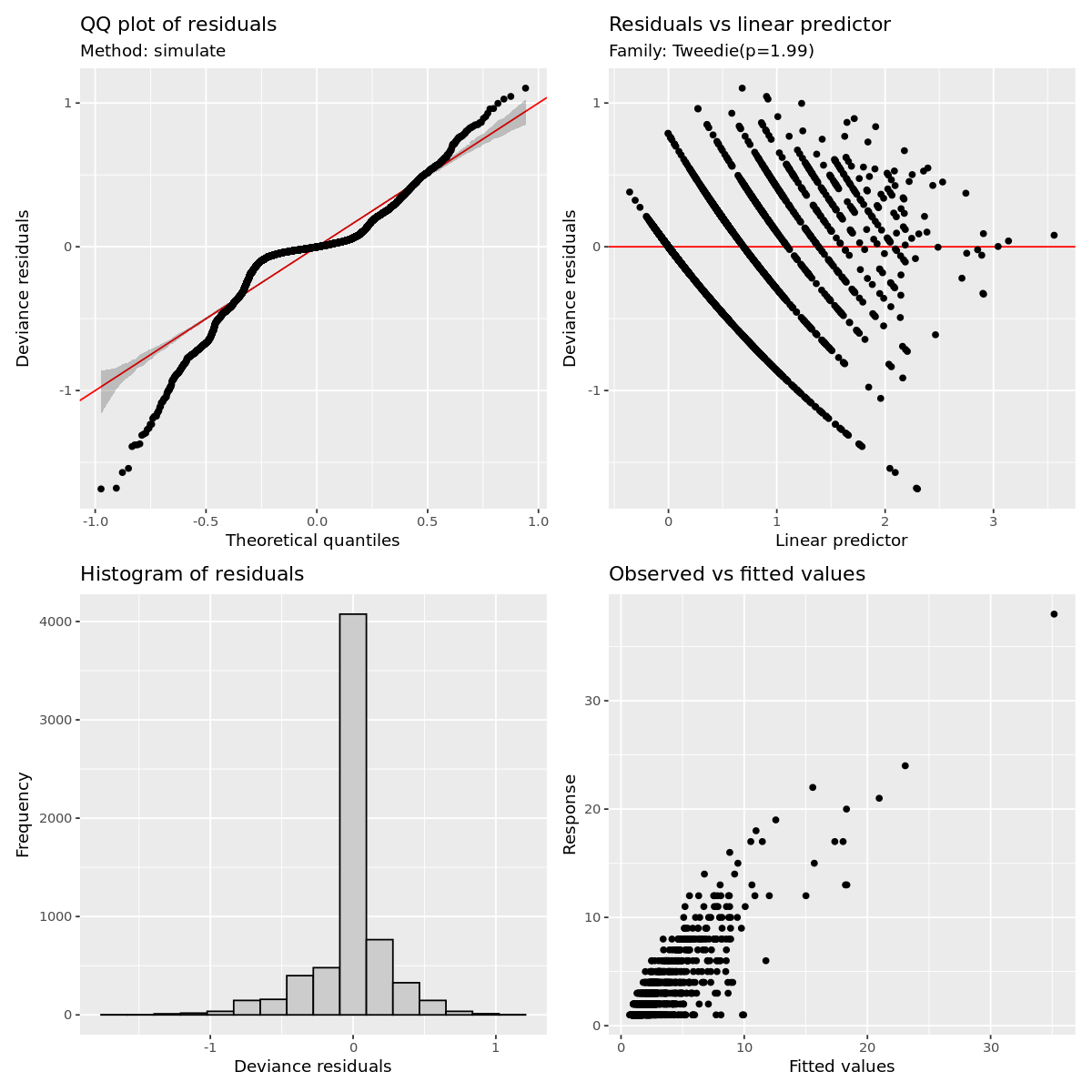
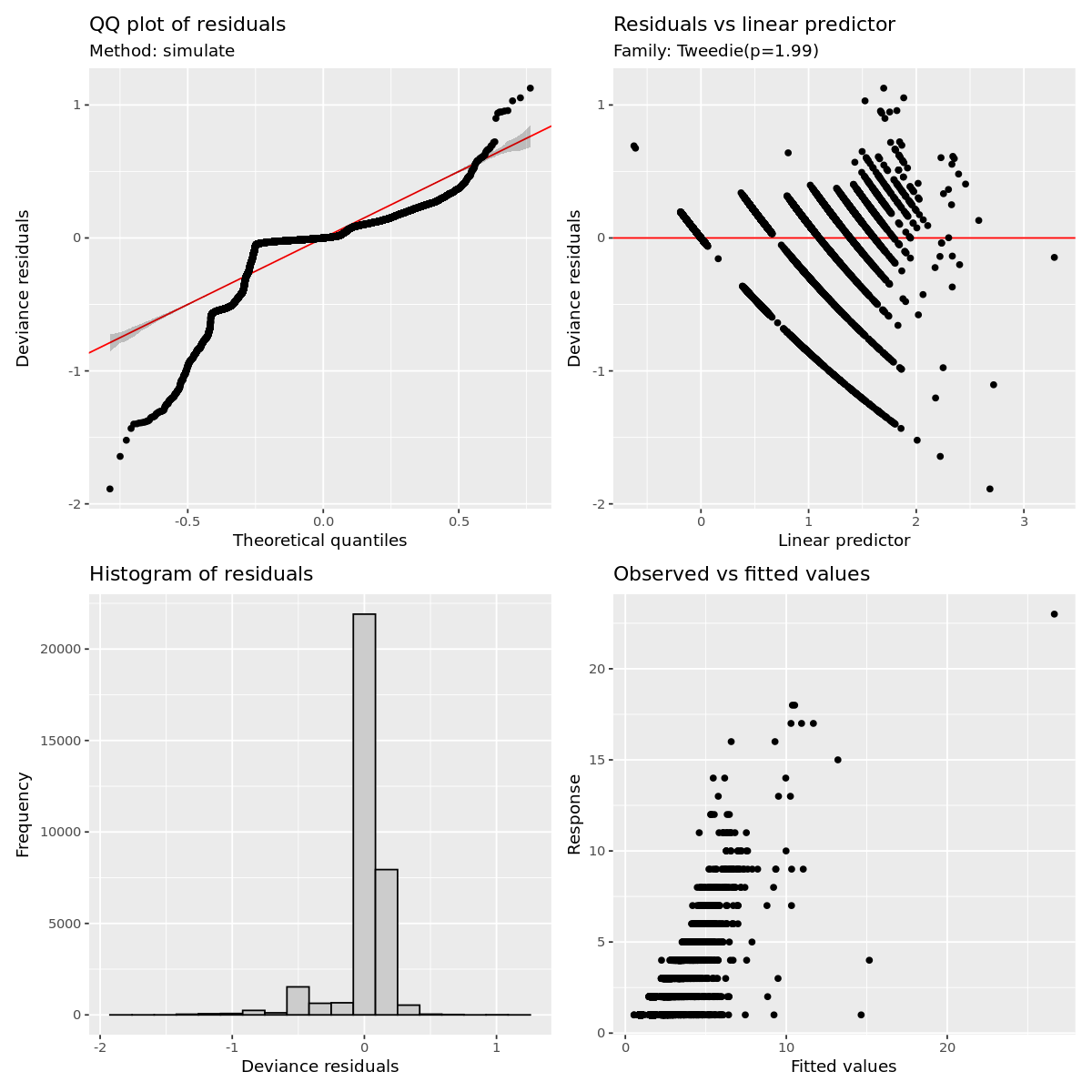
gratia::appraise(outbreak_mod)
Quick model visualization
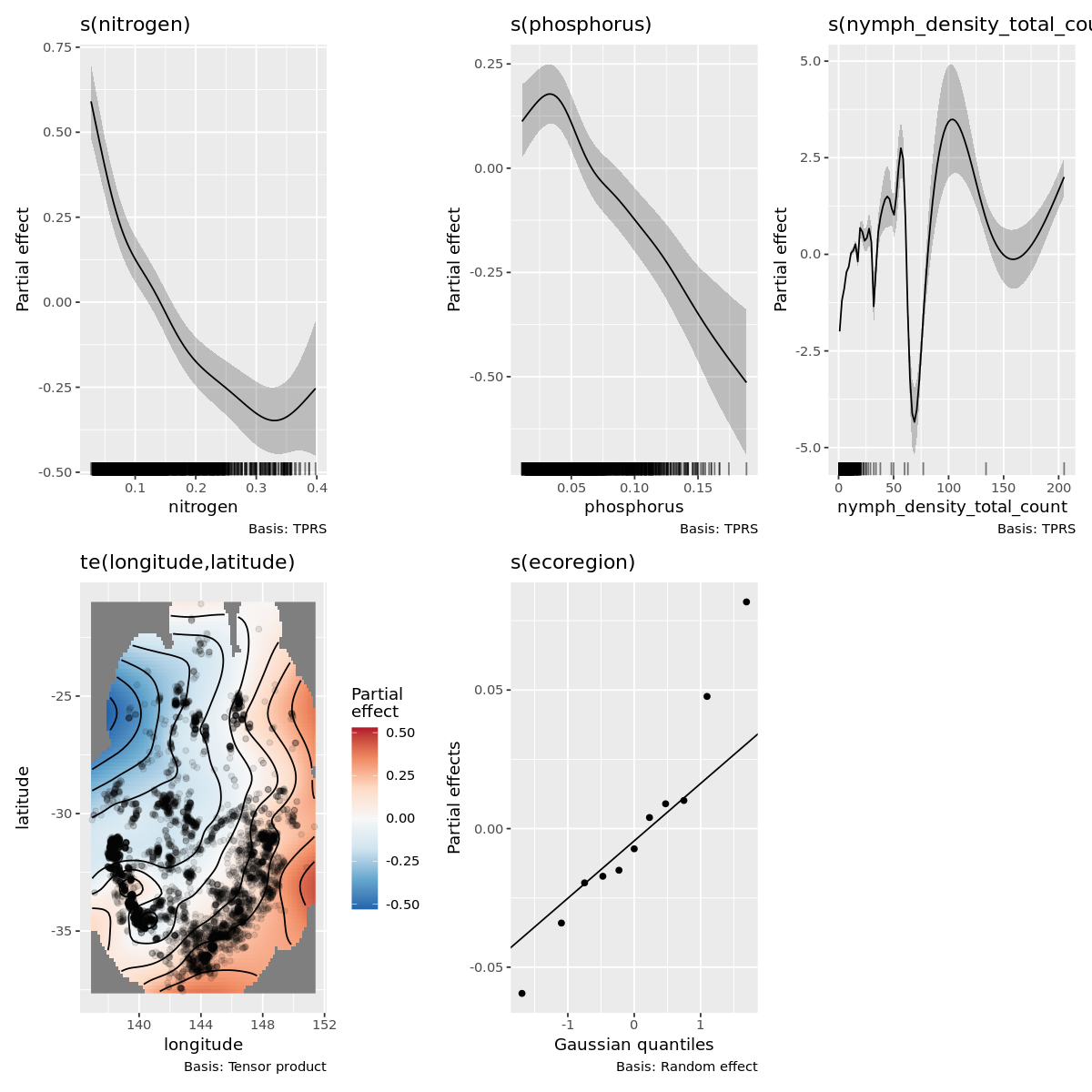
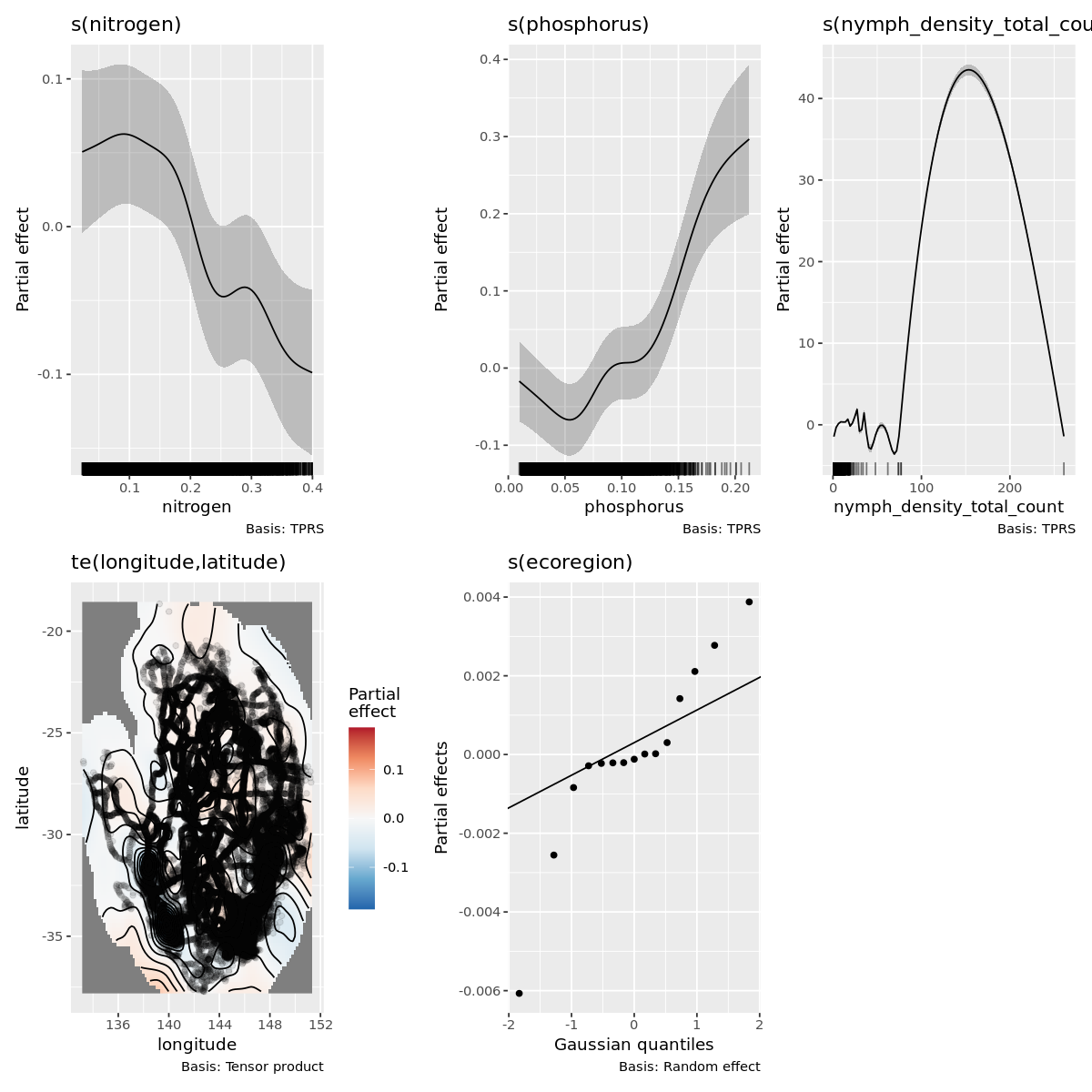
gratia::draw(outbreak_mod)
Outbreak modeling evaluation
Its obvious this model has some fitting issues, espicially with lower values of the dependent variable.
This is likely correlational issues between all the variable and it can muddy the interpretation as well as the number of 1 off observations.
I have already filtered out extreme grid cells that have more than 50 obersvations and that did improve the fit more.
saveRDS(outbreak_mod, here("output/spatial_modeling/model_objects/locust_outbreak_model.rds"))
outbreak_model_smooth_estimates <- smooth_estimates(outbreak_mod,dist=0.1)
write.csv(outbreak_model_smooth_estimates, here("output/spatial_modeling/outbreak_model_smooth_estimates.csv"),row.names=FALSE)
outbreak_model_smooth_estimates <- smooth_estimates(outbreak_mod,dist=0.1)
outbreak_model_smooth_estimates
A smooth_estimates: 10315 × 11
| <chr> |
<chr> |
<chr> |
<dbl> |
<dbl> |
<dbl> |
<dbl> |
<dbl> |
<dbl> |
<dbl> |
<fct> |
| s(nitrogen) |
TPRS |
NA |
0.05056327 |
0.02819440 |
0.02262577 |
NA |
NA |
NA |
NA |
NA |
| s(nitrogen) |
TPRS |
NA |
0.05127533 |
0.02774333 |
0.02643393 |
NA |
NA |
NA |
NA |
NA |
| s(nitrogen) |
TPRS |
NA |
0.05198999 |
0.02732688 |
0.03024208 |
NA |
NA |
NA |
NA |
NA |
| s(nitrogen) |
TPRS |
NA |
0.05271331 |
0.02694706 |
0.03405024 |
NA |
NA |
NA |
NA |
NA |
| s(nitrogen) |
TPRS |
NA |
0.05345310 |
0.02660461 |
0.03785840 |
NA |
NA |
NA |
NA |
NA |
| ⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
| s(ecoregion) |
Random effect |
NA |
-0.0002120522 |
0.005155784 |
NA |
NA |
NA |
NA |
NA |
Naracoorte woodlands |
| s(ecoregion) |
Random effect |
NA |
-0.0002261551 |
0.004471238 |
NA |
NA |
NA |
NA |
NA |
Simpson desert |
| s(ecoregion) |
Random effect |
NA |
0.0014202656 |
0.004640904 |
NA |
NA |
NA |
NA |
NA |
Southeast Australia temperate forests |
| s(ecoregion) |
Random effect |
NA |
0.0038769919 |
0.003721848 |
NA |
NA |
NA |
NA |
NA |
Southeast Australia temperate savanna |
| s(ecoregion) |
Random effect |
NA |
0.0003027099 |
0.004446608 |
NA |
NA |
NA |
NA |
NA |
Tirari-Sturt stony desert |
section 3: nil-observatiom modeling
nil_data <- read_csv(here('data/processed/spatial_modeling/spatial_modeling_locust_nil_model_data.csv'),
show_col_types = FALSE) |>
mutate(ecoregion = factor(ecoregion),nymph_density_count=as.integer(nymph_density_count)) |>
#Filtering out extreme values
filter(nymph_density_count <= 50 & nitrogen <= 0.4) |>
drop_na()
nil_data
A tibble: 33821 × 10
| <dbl> |
<dbl> |
<dbl> |
<dbl> |
<int> |
<dbl> |
<fct> |
<dbl> |
<dbl> |
<dbl> |
| 7280 |
133.4553 |
-27.06065 |
0 |
1 |
1 |
Central Ranges xeric scrub |
208 |
0.03300749 |
0.02376515 |
| 8026 |
133.2392 |
-26.40242 |
0 |
1 |
1 |
Central Ranges xeric scrub |
225 |
0.03097755 |
0.01831567 |
| 8097 |
133.2565 |
-26.49929 |
0 |
1 |
1 |
Central Ranges xeric scrub |
223 |
0.03094262 |
0.01980745 |
| 13279 |
133.7368 |
-27.45913 |
0 |
1 |
1 |
Great Victoria desert |
190 |
0.03268851 |
0.02072279 |
| 13512 |
133.8018 |
-27.53024 |
0 |
2 |
2 |
Tirari-Sturt stony desert |
191 |
0.03296471 |
0.01975296 |
| ⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
| 2099638 |
151.0890 |
-27.07120 |
0 |
1 |
1 |
Brigalow tropical savanna |
647 |
0.1090968 |
0.05359124 |
| 2100024 |
151.1908 |
-27.21150 |
0 |
1 |
1 |
Brigalow tropical savanna |
650 |
0.1135198 |
0.06566815 |
| 2100282 |
151.2569 |
-27.43300 |
0 |
1 |
1 |
Brigalow tropical savanna |
628 |
0.1068338 |
0.06458026 |
| 2101656 |
151.1922 |
-26.90387 |
0 |
1 |
1 |
Brigalow tropical savanna |
637 |
0.1206505 |
0.07311647 |
| 2102441 |
150.9997 |
-26.65369 |
0 |
1 |
1 |
Brigalow tropical savanna |
627 |
0.1157181 |
0.03929814 |
aus <- ne_states(country = 'Australia')
map_dat <- nil_data |>
select(polygon_id,latitude,longitude,nymph_density_count) |>
st_as_sf(coords = c('longitude','latitude'),
crs= "+proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0")
nil_point_map <- aus |>
ggplot() +
geom_sf() +
geom_sf(data=map_dat) +
theme_void()
nil_point_map
nto_plot <- nil_data |>
ggplot(aes(x=nitrogen,y=nymph_density_count)) +
geom_point(pch=21,alpha=0.3,size=0.6) +
geom_smooth() +
theme_pubr()
pto_plot <- nil_data |>
ggplot(aes(x=phosphorus,y=nymph_density_count)) +
geom_point(pch=21,alpha=0.3,size=0.6) +
geom_smooth() +
theme_pubr()
nto_plot + pto_plot
`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
nil_mod <- bam(nymph_density_count ~
s(nitrogen,k=10,bs='tp') +
s(phosphorus,k=10,bs='tp') +
s(nymph_density_total_count,k=25,bs='tp') +
te(longitude,latitude,bs=c('gp','gp'),k=25) +
s(ecoregion,bs='re'),
select = TRUE,
discrete = TRUE,
nthreads = 20,
family = tw(),
data = nil_data)
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
A matrix: 5 × 4 of type dbl
| s(nitrogen) |
9 |
6.426332 |
0.9719328 |
0.0425 |
| s(phosphorus) |
9 |
6.406880 |
0.9658401 |
0.0150 |
| s(nymph_density_total_count) |
24 |
22.408362 |
0.9912682 |
0.3475 |
| te(longitude,latitude) |
624 |
131.476046 |
0.9515954 |
0.0050 |
| s(ecoregion) |
15 |
2.726108 |
NA |
NA |
concurvity(nil_mod,full=FALSE)
- $worst
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.8307967 |
0.8685621 |
0.9974470 |
1.0000000 |
1.0000000 |
| s(nitrogen) |
0.8307967 |
1.0000000 |
0.9004807 |
0.8637565 |
0.8800706 |
0.8556794 |
| s(phosphorus) |
0.8685621 |
0.9004807 |
1.0000000 |
0.9113526 |
0.8792777 |
0.8697074 |
| s(nymph_density_total_count) |
0.9974470 |
0.8637565 |
0.9113526 |
1.0000000 |
0.9975198 |
0.9974679 |
| te(longitude,latitude) |
1.0000000 |
0.8800708 |
0.8792781 |
0.9975198 |
1.0000000 |
1.0000000 |
| s(ecoregion) |
1.0000000 |
0.8556794 |
0.8697074 |
0.9974679 |
1.0000000 |
1.0000000 |
- $observed
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.7868098 |
0.8083066 |
0.8551952 |
0.004546396 |
0.06430443 |
| s(nitrogen) |
0.8307967 |
1.0000000 |
0.8181989 |
0.8397272 |
0.044026705 |
0.14027235 |
| s(phosphorus) |
0.8685621 |
0.8614793 |
1.0000000 |
0.9002601 |
0.061211576 |
0.17528098 |
| s(nymph_density_total_count) |
0.9974470 |
0.8241006 |
0.8383076 |
1.0000000 |
0.026668080 |
0.07966450 |
| te(longitude,latitude) |
1.0000000 |
0.8215499 |
0.8199754 |
0.8727459 |
1.000000000 |
0.81144707 |
| s(ecoregion) |
1.0000000 |
0.8028665 |
0.8106659 |
0.8580396 |
0.381904995 |
1.00000000 |
- $estimate
-
A matrix: 6 × 6 of type dbl
| para |
1.0000000 |
0.6918896 |
0.7004913 |
0.9870538 |
0.002128188 |
0.1470001 |
| s(nitrogen) |
0.8307967 |
1.0000000 |
0.8371775 |
0.8351752 |
0.008066931 |
0.2057451 |
| s(phosphorus) |
0.8685621 |
0.8295907 |
1.0000000 |
0.8755378 |
0.003650041 |
0.1498173 |
| s(nymph_density_total_count) |
0.9974470 |
0.7624890 |
0.8394230 |
1.0000000 |
0.003104366 |
0.1504301 |
| te(longitude,latitude) |
1.0000000 |
0.8375298 |
0.7564592 |
0.9879833 |
1.000000000 |
0.8873811 |
| s(ecoregion) |
1.0000000 |
0.7927771 |
0.7150555 |
0.9872542 |
0.021132004 |
1.0000000 |
gratia::appraise(nil_mod)
Family: Tweedie(p=1.99)
Link function: log
Formula:
nymph_density_count ~ s(nitrogen, k = 10, bs = "tp") + s(phosphorus,
k = 10, bs = "tp") + s(nymph_density_total_count, k = 25,
bs = "tp") + te(longitude, latitude, bs = c("gp", "gp"),
k = 25) + s(ecoregion, bs = "re")
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.43919 0.02338 61.55 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(nitrogen) 6.426 9 35.340 <2e-16 ***
s(phosphorus) 6.407 9 28.867 <2e-16 ***
s(nymph_density_total_count) 22.408 24 3199.357 <2e-16 ***
te(longitude,latitude) 131.476 557 3.302 <2e-16 ***
s(ecoregion) 2.726 14 0.361 0.0352 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
R-sq.(adj) = 0.787 Deviance explained = 87.9%
fREML = -8209.3 Scale est. = 0.035064 n = 33821
Quick modeling visualization
saveRDS(nil_mod, here("output/spatial_modeling/model_objects/locust_nil_model.rds"))
outbreak_model_smooth_estimates <- smooth_estimates(nil_mod,dist=0.1)
write.csv(outbreak_model_smooth_estimates, here("output/spatial_modeling/nil_model_smooth_estimates.csv"),row.names=FALSE)
Section 4 - Locust outbreaks and mean annual preciptation modeling
A tibble: 33969 × 10
| <dbl> |
<dbl> |
<dbl> |
<dbl> |
<int> |
<dbl> |
<fct> |
<dbl> |
<dbl> |
<dbl> |
| 7280 |
133.4553 |
-27.06065 |
0 |
1 |
1 |
Central Ranges xeric scrub |
208 |
0.03300749 |
0.02376515 |
| 8026 |
133.2392 |
-26.40242 |
0 |
1 |
1 |
Central Ranges xeric scrub |
225 |
0.03097755 |
0.01831567 |
| 8097 |
133.2565 |
-26.49929 |
0 |
1 |
1 |
Central Ranges xeric scrub |
223 |
0.03094262 |
0.01980745 |
| 13279 |
133.7368 |
-27.45913 |
0 |
1 |
1 |
Great Victoria desert |
190 |
0.03268851 |
0.02072279 |
| 13512 |
133.8018 |
-27.53024 |
0 |
2 |
2 |
Tirari-Sturt stony desert |
191 |
0.03296471 |
0.01975296 |
| ⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
⋮ |
| 2099638 |
151.0890 |
-27.07120 |
0 |
1 |
1 |
Brigalow tropical savanna |
647 |
0.1090968 |
0.05359124 |
| 2100024 |
151.1908 |
-27.21150 |
0 |
1 |
1 |
Brigalow tropical savanna |
650 |
0.1135198 |
0.06566815 |
| 2100282 |
151.2569 |
-27.43300 |
0 |
1 |
1 |
Brigalow tropical savanna |
628 |
0.1068338 |
0.06458026 |
| 2101656 |
151.1922 |
-26.90387 |
0 |
1 |
1 |
Brigalow tropical savanna |
637 |
0.1206505 |
0.07311647 |
| 2102441 |
150.9997 |
-26.65369 |
0 |
1 |
1 |
Brigalow tropical savanna |
627 |
0.1157181 |
0.03929814 |
outbreak_map_mod <- bam(nymph_density_count ~
s(bio12,bs='tp',k=10) +
s(nymph_density_total_count,k=25,bs='tp') +
te(longitude,latitude,bs=c('gp','gp'),k=25) +
s(ecoregion,bs='re'),
select = TRUE,
discrete = TRUE,
nthreads = 20,
family = tw(),
data = outbreak_data)
nil_map_mod <- bam(nymph_density_count ~
s(bio12,bs='tp',k=10) +
s(nymph_density_total_count,k=25,bs='tp') +
te(longitude,latitude,bs=c('gp','gp'),k=25) +
s(ecoregion,bs='re'),
select = TRUE,
discrete = TRUE,
nthreads = 20,
family = tw(),
data = nil_data)
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
Warning message in estimate.theta(theta, family, y, mu, scale = scale1, wt = G$w, :
“step failure in theta estimation”
saveRDS(outbreak_map_mod, here("output/spatial_modeling/model_objects/locust_outbreak_map_model.rds"))
saveRDS(nil_map_mod, here("output/spatial_modeling/model_objects/locust_nil_map_model.rds"))
map_outbreak_model_smooth_estimates <- smooth_estimates(outbreak_map_mod,dist=0.1)
map_nill_model_smooth_estimates <- smooth_estimates(nil_map_mod,dist=0.1)
write.csv(map_outbreak_model_smooth_estimates, here("output/spatial_modeling/map_outbreak_model_smooth_estimates.csv"),row.names=FALSE)
write.csv(map_nill_model_smooth_estimates, here("output/spatial_modeling/map_nill_model_smooth_estimates.csv"),row.names=FALSE)